Login
You are currently not logged in! Enter your authentication credentials below to log in. You need to have cookies enabled to log in.
Find structure element
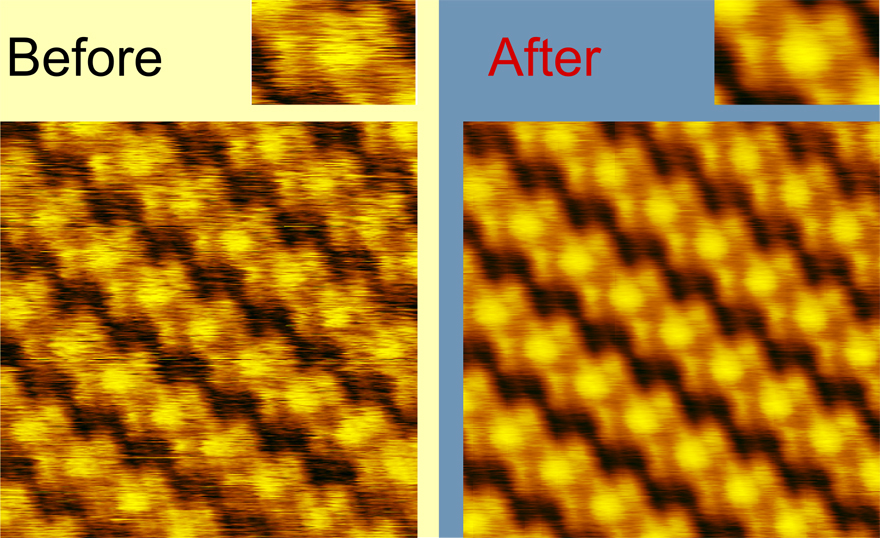
It is not rare in scanning probe microscopy and especially in scanning tunnel microscopy that scientists work with highly organized organic films or with cristal structures. Objects of this type are suited to microscopy investigation on molecular level, the images may look spectacular and in some cases it is possible to build the model of cristal or film unit cell. The common problem of images of this type is their noisiness. Find structure element function allows to build averaged unit cell image or to improve the original image by means of correlation analysis.
The order of work
Open an image of an ordered structure you want to process. First of all select the fragment with minimal noise and artifacts on which one or several unit cells are presented, duplicate this fragment. Return to the initial image, it must be active when you call the function. Call Find structure element from Mathematics menu → Correlation.
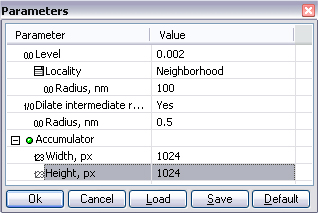
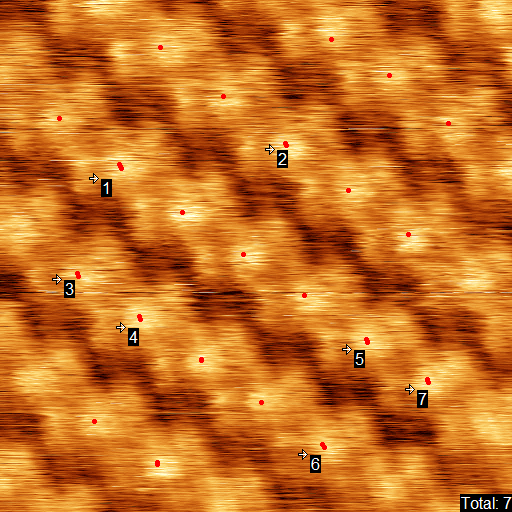
Select the copied element in the appeared window. Two more windows will appear - parameters window and a window with the resulting image. The function detaches structure elements by determination of local maximums of cross correlation between two images. The points of the maximums are marked with red dots on the initial image and our intention here is to achieve the correct determination of all the maximums by adjusting the parameters of the function. The rigidity in maximums picking may be different, it is defined by Level parameter. this parameter varies between 0 and 1, 0 corresponds to the most strict picking of maximums and often gives a good result. Normally the best result may be achieved with small values of this parameter. Select a string with the Level parameter and find the optimal value by clicking arrows near it.
The size of the area in which maximums are searched may coincide with the size of structure element image used for correlation, in this case the value of Locality parameter mast be set to Structure element. It is reasonable if you have selected one or few unit cells for cross correlation. If you have isolated an area consisting of several unit cells, then select Neighborhood and specify the size of the neighborhood in nanometers. After selecting the two options described above, some of the centers of elementary cells may be identified twice by mistake:
In this case set the value Yes to the parameter Dilate intermediate results to merge closely located maximums. The radius of merging is defined by Radius, nm parameter.
A groupe Accumulator integreate the parameters of resulting image size. One can set for the final image the same XY sizes as initial image has, in this case the new filtered full image will be formed. One can also form up a picture with a part of initial image only, for example an image of one unit cell.
Examples
In general, this feature provides a very powerful tool to improve the images of periodic structures with molecular and atomic resolution. But it requires fine-tuning of the parameters, and if you do not get the desirable result with your images, see the section Examples, where details on the parameter values used in different situations will be given .